Perpetuation of Gene Expression Patterns
Phenomena such as paramuation and RNA interference (RNAi) have revealed that changes in gene regulation can persist for many generations as part of reconfigured cell codes. Such transgenerational gene silencing can be initiated in C. elegans using double-stranded RNA (dsRNA) or a class of small RNAs called piRNAs. We recently discovered another phenomenon whereby mating can reliably initiate transgenerational gene silencing. These different ways of initiating transgenerational gene silencing provide opportunities to understand how molecules that can change the cell code gain access to the germline and how silencing is transmitted across generations as part of a reconfigured cell code.
Initiation by the entry of double-stranded RNA into the germline: The somatic cells that generate our body are separated during early development from the germ cells that make sperm or egg. Therefore, any molecules that can change gene regulatory information need to gain access to the germline to cause transgenerational effects. Early experiments using the injection of dsRNA or feeding worms bacteria that express dsRNA suggested that dsRNA and/or molecules derived from it could be transported to the germline in C. elegans.
Injection: To directly visualize the fate of extracellular dsRNAs, we injected fluorescently labeled 50-bp dsRNA into the body cavity that surrounds all tissues, including the germline, in C. elegans. We found that dsRNA entered oocytes along with yolk and reached progeny (Marré et al., 2016). Surprisingly, we found that such delivery of extracellular RNA from parent to progeny did not require entry into the cytosol within the parent. Entry into the cytosol can subseqeuntly occur in progeny through the dsRNA importer SID-1 - a conserved protein with homologs in many animals, including humans. 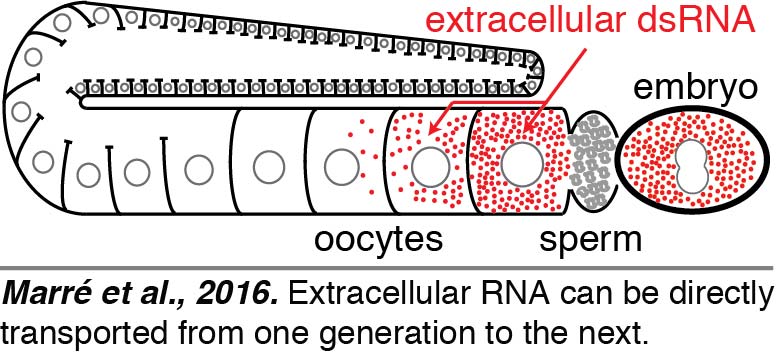 Thus, the dsRNA that enters oocytes along with yolk can presumably be held within intracellular vesicles and reach embryos. These results raise the intriguing possibility that extracellular RNAs that are secreted – potentially in response to changes in a parent – can directly reach progeny and regulate gene expression. However, other studies suggest additional routes for the transport of dsRNA into germ cells that remain to be elucidated.
Thus, the dsRNA that enters oocytes along with yolk can presumably be held within intracellular vesicles and reach embryos. These results raise the intriguing possibility that extracellular RNAs that are secreted – potentially in response to changes in a parent – can directly reach progeny and regulate gene expression. However, other studies suggest additional routes for the transport of dsRNA into germ cells that remain to be elucidated.
Feeding: A robust and easy way to initiate RNA silencing is feeding worms bacteria that express dsRNA (feeding RNAi). 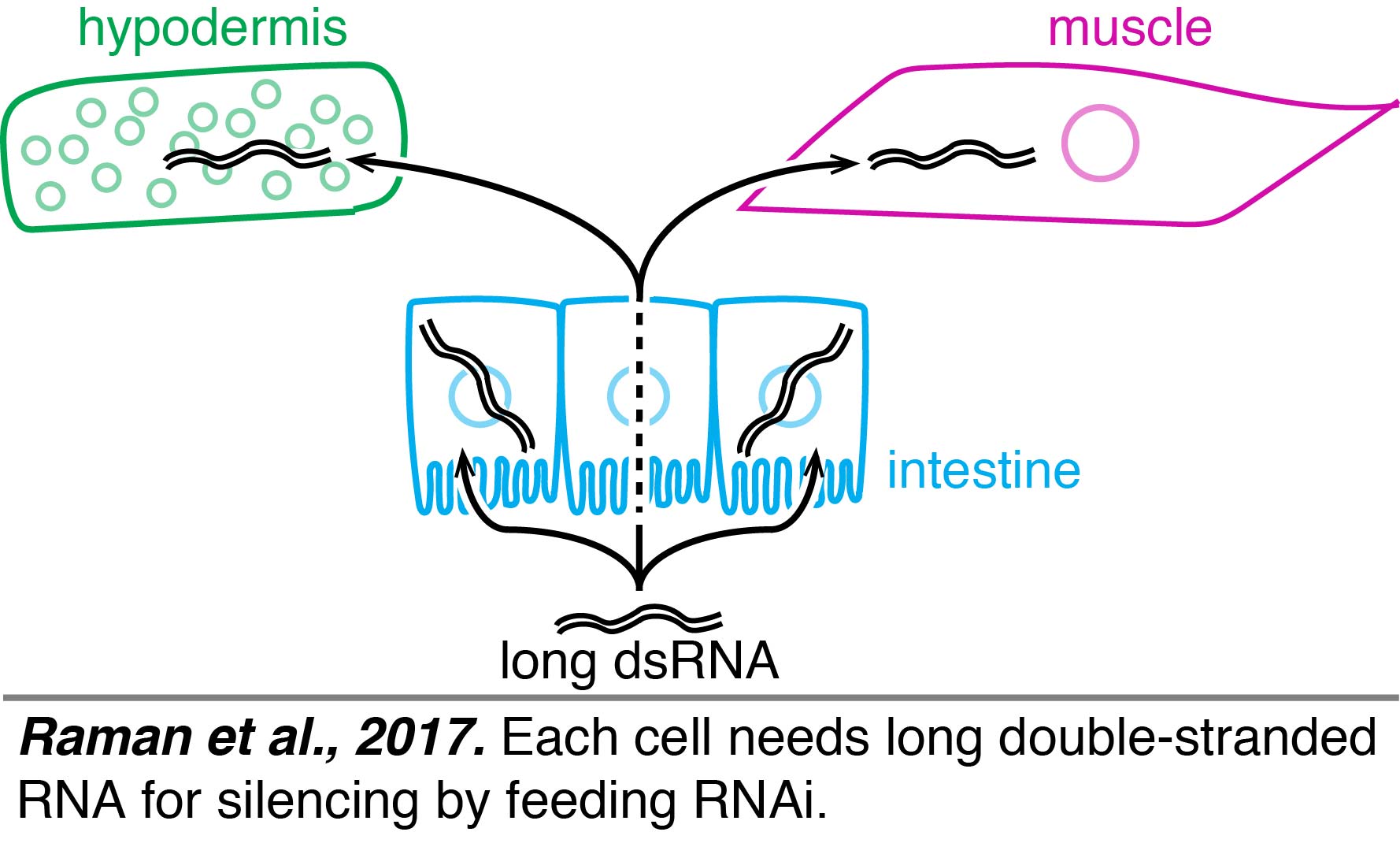 Using feeding RNAi, multiple groups have contributed to an understanding of how long dsRNA is processed into shorter dsRNAs and used for finding matching sequences to cause gene silencing. One way to infer what RNAs need to enter each tissue to cause silencing is by restricting the processing of long dsRNA to specific tissues. Such tissue-specific rescue experiments using single-copy transgenes suggest that each cell needs long dsRNA for silencing by feeding RNAi (Raman et al., 2017).
Expression: Recent work in many organisms is revealing that cells can communicate with each other through the direct transport of macromolecules (e.g. RNA). Secreted vesicles with such macromolecules that are found in human blood are being hotly pursued as valuable diagnostic markers for the health of internal organs. However, the mechanisms that enable this mode of cell communication are not well understood.
We found that dsRNA expressed within neurons could enter the germline and cause transgenerational gene silencing (Devanapally et al., 2015).
Using feeding RNAi, multiple groups have contributed to an understanding of how long dsRNA is processed into shorter dsRNAs and used for finding matching sequences to cause gene silencing. One way to infer what RNAs need to enter each tissue to cause silencing is by restricting the processing of long dsRNA to specific tissues. Such tissue-specific rescue experiments using single-copy transgenes suggest that each cell needs long dsRNA for silencing by feeding RNAi (Raman et al., 2017).
Expression: Recent work in many organisms is revealing that cells can communicate with each other through the direct transport of macromolecules (e.g. RNA). Secreted vesicles with such macromolecules that are found in human blood are being hotly pursued as valuable diagnostic markers for the health of internal organs. However, the mechanisms that enable this mode of cell communication are not well understood.
We found that dsRNA expressed within neurons could enter the germline and cause transgenerational gene silencing (Devanapally et al., 2015). 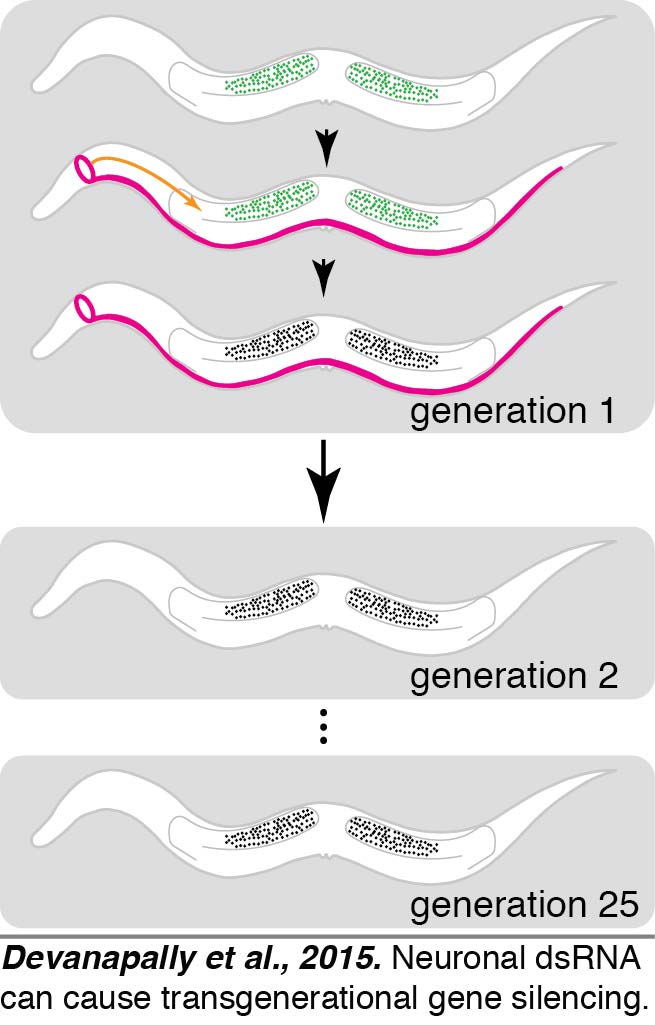 C. elegans neurons (magenta) can send gene-specific messages in the form of dsRNA to the germline (green) and cause transgenerational gene silencing that can last for more than 25 generations. Neuronal dsRNAs enter germ cells through SID-1. These results can have profound implications for our understanding of evolution and behavior. We have begun a UMD-funded program called the Transgenerational Brain Initiative (http://www.fire.umd.edu/streams-TBI.html) that trains ~35 undergraduates each year in original research aimed at discovering which neuron(s) are the best exporters of dsRNA to the germline. Current models for the transgenerational inheritance of silencing in C. elegans propose reinforcement through the production of small RNAs and associated chromatin modifications in each generation. Most aspects of this model, however, remain to be tested and only a few protein factors required for this process are known.
C. elegans neurons (magenta) can send gene-specific messages in the form of dsRNA to the germline (green) and cause transgenerational gene silencing that can last for more than 25 generations. Neuronal dsRNAs enter germ cells through SID-1. These results can have profound implications for our understanding of evolution and behavior. We have begun a UMD-funded program called the Transgenerational Brain Initiative (http://www.fire.umd.edu/streams-TBI.html) that trains ~35 undergraduates each year in original research aimed at discovering which neuron(s) are the best exporters of dsRNA to the germline. Current models for the transgenerational inheritance of silencing in C. elegans propose reinforcement through the production of small RNAs and associated chromatin modifications in each generation. Most aspects of this model, however, remain to be tested and only a few protein factors required for this process are known.
Despite the progress in understanding how extracellular dsRNA causes silencing within the germline, many questions remain: (1) What is the full complement of routes taken by dsRNA from one generation to the next? (2) How is dsRNA exported from cells? (3) What RNAs transcribed from the C. elegans genome or from ingested bacteria reach the germline? (4) What other gene regulatory molecules are transported across generations?
Initiation by mating: The recent ability to precisely modify the C. elegans genome led several groups 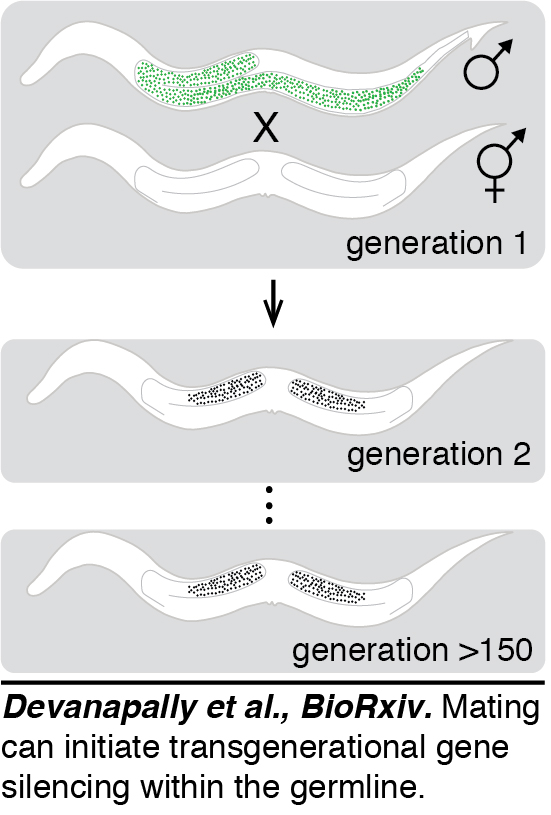 to uncover a transgenerational gene silencing response that can act on single-copy transgenes. This silencing resembles paramutation and was shown to depend on a class of small RNAs called piRNAs. We found a single-copy transgene that shows stable expression but that can be silenced simply by mating males with the transgene to hermaphrodites that lack the transgene - a phenomenon we named mating-induced silencing (Devanapally et al., BioRxiv). This silencing can be stable for >150 generations. Genetic requirements for this phenomenon suggest that initiation of silencing requires piRNAs and maintenance of silencing requires the production of additional small RNAs potentially associated with chromatin modifications in each generation. Because mating is an easy approach to reliably initiate transgenerational silencing, it can be coupled with CRISPR-based genetic and epigenetic engineering approaches to uncover the features that determine transgenerationally stable expression versus silencing and the switch between these two configurations of the cell code.
Our future efforts will address the following questions:
(1) What features of a transgenic locus determine susceptibility to mating-induced silencing?
(2) Are there endogenous loci that are susceptible to mating-induced silencing?
(3) What regulatory mechanisms determine whether a gene is expressed or silenced?
(4) How can such mechanisms be changed to reconfigure expression of a gene for many generations?
to uncover a transgenerational gene silencing response that can act on single-copy transgenes. This silencing resembles paramutation and was shown to depend on a class of small RNAs called piRNAs. We found a single-copy transgene that shows stable expression but that can be silenced simply by mating males with the transgene to hermaphrodites that lack the transgene - a phenomenon we named mating-induced silencing (Devanapally et al., BioRxiv). This silencing can be stable for >150 generations. Genetic requirements for this phenomenon suggest that initiation of silencing requires piRNAs and maintenance of silencing requires the production of additional small RNAs potentially associated with chromatin modifications in each generation. Because mating is an easy approach to reliably initiate transgenerational silencing, it can be coupled with CRISPR-based genetic and epigenetic engineering approaches to uncover the features that determine transgenerationally stable expression versus silencing and the switch between these two configurations of the cell code.
Our future efforts will address the following questions:
(1) What features of a transgenic locus determine susceptibility to mating-induced silencing?
(2) Are there endogenous loci that are susceptible to mating-induced silencing?
(3) What regulatory mechanisms determine whether a gene is expressed or silenced?
(4) How can such mechanisms be changed to reconfigure expression of a gene for many generations?
Back to research
Last updated: Feb 2018
Web Accessibility